AutoBioML: Building a Generalist Multi-Agent Framework for BioML
With solution to Agentic BioML Challenge #1

Last month I launched the Agentic BioML Challenge, a series of six tasks designed to test whether large-language-model (LLM) agents can solve real-world biomedical machine learning (BioML) problems without task-specific code or rigid workflows. For every task, the agents must operate as generalists and are evaluated exclusively on held-out data they have never encountered.
The first task involved constructing an “epigenetic clock” (age-predictor) based on DNA-methylation (DNAm) data. Agents received the AltumAge methylation compendium (Camillo 2022) which has matched patient ages included, and were tasked with building a model that meets two performance targets: Pearson r > 0.9 and mean-absolute error (MAE) < 10 years. Although this is a well-understood problem, our exercise evaluated whether a generalist multi-agent system could autonomously solve this BioML challenge end-to-end.
To provide feedback without compromising the final assessment, the held-out evaluation dataset was split into a public and a private test set (splits are based on the study that data originates from, not sample/donor). Agents could evaluate performance on the public subset during development to identify obvious preprocessing or modelling errors; final performance was calculated on the hidden private subset. This mirrors the “public / private leaderboard” structure used in Kaggle competitions and allowed me to quantify the value of interim feedback for the agent system.
Here’s how our experiments went:
1️⃣ Data at a Glance
Total samples: 13,504
Agent-accessible training set: 10,620 samples
Held-out test set: 2,884 samples split into
Public subset: 1,262 samples (15 studies)
Private subset: 1,622 samples (15 studies)
The training set spanned 64 tissue types (whole blood was most common), ages –1 → 114 yr (median ≈ 47 yr) and a near-even gender balance. Public and private subsets follow similar distributions but had different studies, so the model had to generalize.
2️⃣ The AutoBioML Framework
To solve the task, I introduced AutoBioML, a multi-agent BioML generalist framework. At a high level, AutoBioML:
Mimics the dynamics of a BioML research team with different laboratory agent personas (e.g., “Bioinformatics Expert”) that work together collaboratively.
Uses a planning-implementation loop with autonomous code writing/execution to enable agents to design and run experiments.
Provides tools for agents to interact with files, search literature, and even read and review PNG plots.
The code for AutoBioML is open-source and contributions are welcome: https://github.com/AIxBio/AgBioML-Challenge. Here is a detailed description:
🧑🔬 Agent Teams & Roles
Planning Team
Principal Scientist – steers strategy, arbitrates disagreements, searches for relevant literature, enforces documentation discipline.
Bioinformatics Expert – recommends preprocessing approaches and identifies best-practices for biodata handling from literature.
ML Expert – surveys state-of-the-art models from literature, drafts the training plan, compares options against published results.
Engineering team
Implementation engineer - Executes the spec from planning team with a ReAct loop (Thought → Action → Observation). Uses a dockerized environment for code execution. Installs packages as needed.
Data Science Critic - Reviews code, metrics, and plots after each engineer hand-off. Can veto a run for: brittle pipelines, resource under-use, violated rules, or undocumented choices.
🔁 Workflow
Planning – round-robin discussion amongst the planning team; outputs an implementation spec for engineering team to follow.
Engineering – engineer team executes specification. Their actions typically involve performing EDA, building models, running tests on public test set (if this option is provided), iterating as needed. Critic writes final summary of implementation to be sent to the planning team.
Iteration - Repeat step 1 and 2 up to 25 times.
Completion – once satisfied with the project, the principal scientist signs off (stamps “TASK_COMPLETE”), triggers final private-test evaluation.
🔧 Built-In Agent Tools
Research – Perplexity Search, Webpage Parser.
Data IO – Arrow/Feather readers, directory search, safe file writes.
Visualization – Plot Analyzer (GPT-4o-mini looks at figures, summarizes their findings, and flags issues with them).
Evaluation –
evaluate_on_public_test()returns public test-set eval metrics so agents can course-correct.
🏁 Model Evaluations
Test set is split 50 % public / 50 % private.
Agents can score on the public test set first (if enabled).
When satisfied with the model performance, they output
predictions.arrowfrom inference on the private test set; the framework then runs the evaluation script, generating final metrics.
3️⃣ Challenge #1 Solution (GPT-4.1 + Public Eval)
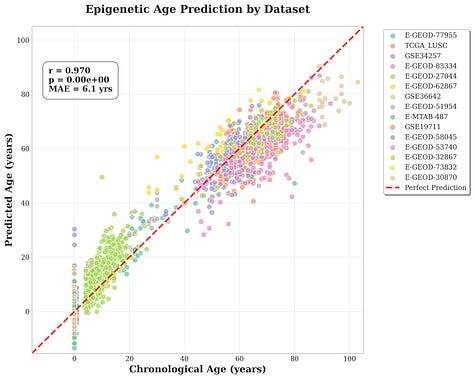
Private-test metrics
Pearson r = 0.97
MAE = 6.10 yr
RMSE = 7.81 yr
R² = 0.933
Criteria passed ✅
How the agents built this solution:
Agents used Perplexity Search to summarize the epigenetic-clock literature (e.g., Horvath 2013) and learn best practices for DNAm preprocessing.
They settled on a pipeline: quantile normalization → variance filter → selected top 8k CpGs based on correlation with age → ElasticNet with K-fold CV + hyper-parameter grid search.
Agents also tested HistGradientBoostingRegressor but it performed worse than ElasticNet so it wasn’t selected.
Final model trained on full training set.
Public eval was performed and model scored well (Pearson r = 0.946, MAE = 5.55).
All inputs, outputs, agent code, eval results, and agent chat summaries are provided publicly at this link in tar.gz format.
4️⃣ Mini-benchmark of LLM & public eval impact
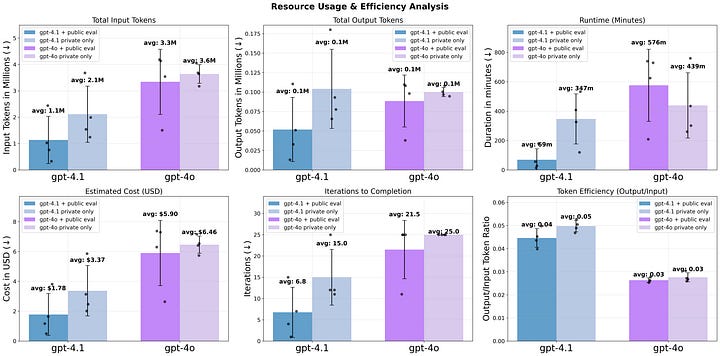
I ran four conditions (four replicates each):
GPT-4.1 + public evaluation
Mean Pearson ≈ 0.969; MAE ≈ 6.0 yr
100 % success; ~7 iterations (planning → engineering team cycles) per run; cheapest per result
GPT-4.1 without public evaluation
Pearson ≈ 0.958; MAE ≈ 11.6 yr
Only 2 of 4 runs met the target, required full 15-iteration budget
GPT-4o without public evaluation
Pearson ≈ 0.880; MAE ≈ 12.0 yr
One successful run but three failures; very token-hungry
GPT-4o + public evaluation
Only one run made it to evaluation (other three ran over 25-iteration limit).

Take-aways
Feedback loops matter: letting agents evaluate models against public test data improves performance and efficiency greatly.
Better model = better performance: GPT-4.1 was steadier and cheaper than GPT-4o.
Efficiency pays off: the best-performing agents converged to a solution in under three hours and <$4 in API costs.
What’s Next — Challenge 02: RSNA Mammography
The clocks are ticking nicely; time for pixels. Challenge 02 tasks the agents with spotting breast cancer in screening mammograms and climbing into the top 25 % of the RSNA kaggle leaderboard.
I’m also planning on tweaking and improving AutoBioML to: (1) simplify task config files, (2) add more sophisticated code-editing tools, and (3) improve agent access to package documentation.
If you’d like to poke the code, reproduce the runs, or propose new tasks, the GitHub is open to you; contributions are very welcome. Let’s keep making 🤖 do science.

